A microbiome is the collective genetic material of the community of microorganisms living in a specific habitat, which can be a niche related to a living organism, e.g., the skin, mouth, or gut, or in the environment, e.g., in soil or water.
Our understanding of the complex relationships between these different microorganisms and the intricate mechanisms by which they influence health and disease has grown tremendously in recent years. Investigating and evaluating an individual’s microbiome and understanding the impact of altering it are important in many areas of medicine.
Microbiomes comprise bacteria, archaea, viruses, and fungi, and the DNA of these microorganisms is studied to gain insights into their diversity, abundance, and functional potential. However, extracting DNA from complex microbiome samples can be challenging due to low DNA yields and the presence of inhibitors and background DNA from the host organism.
This article discusses how to isolate and analyse DNA from three important human microbiomes; the gut, skin, and oral microbiomes.
Human Microbes
The largest human microbiome by mass is the gut microbiome, which weighs approximately 2kg[i] and performs a wide range of functions critical for maintaining health and wellbeing, e.g., aiding digestion, synthesizing essential nutrients, and metabolizing medications. Imbalances in the gut microbiome, known as dysbiosis, are associated with many health conditions, including chronic diseases[ii], cancers[iii], mental health disorders[iv], and gastrointestinal disorders[v].
The second largest microbial community in humans is the oral microbiome[vi]. Dysbiosis in the oral microbiome can lead to various oral health issues[vii] and systemic diseases.[viii] [ix] However, most oral microbiota are beneficial, contributing to good oral health by protecting against pathogens and maintaining a healthy pH balance. The composition of the oral microbiome may be a proxy for microbiomes in other niches, including that of the gut and lung.
The skin microbiome has a relatively low biomass, but a healthy skin microbiome is also critical for good health. Interactions between members of the skin microbiota can prevent colonization by pathogenic bacteria[x], and many common skin diseases are associated with dysbiosis of the skin microbiome[xi].
Collecting DNA for Microbiome Analysis
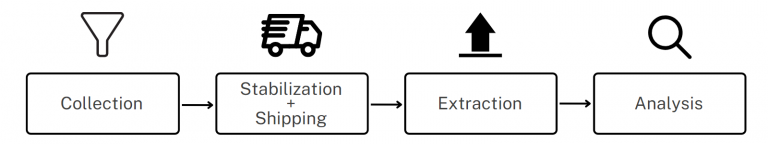
Microbiome composition can vary significantly between individuals and even within the same individual over time. Several factors contribute to success in obtaining a representative sample of DNA for microbiome analysis.
1. Sampling, Shipping, and Storage
Microbiome samples are very sensitive to shipping and storage conditions. Not only does DNA degrade if not stored correctly, but some microorganisms in the sample may continue to grow at the expense of others, meaning that the sample is no longer representative of the original community. Traditional methods of shipping and storing microbiome samples involved freezing, which presents logistical challenges with maintaining a cold chain during transport, and risks sample loss due to freezer failure.
Isohelix has addressed these issues with their range of microbiome DNA collection kits, which include stabilization reagents that instantly stabilize samples, providing an accurate snapshot of the microbiome. Samples can then be shipped and stored at ambient temperatures.
Gut microbiome sample collection can be achieved at home or in the clinic, using rectal swabs or stool samples. The simplest method, as employed by the Isohelix StoolFix Gut Microbiome stabilisation kit (STF), is to simply brush the outside of a stool sample with a swab, before placing the swab into a tube containing stabilisation solution. This is much easier than the traditional “scoop” method of stool sampling.
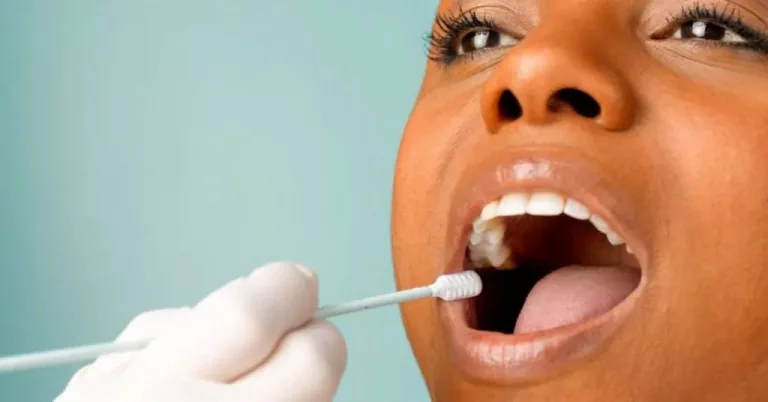
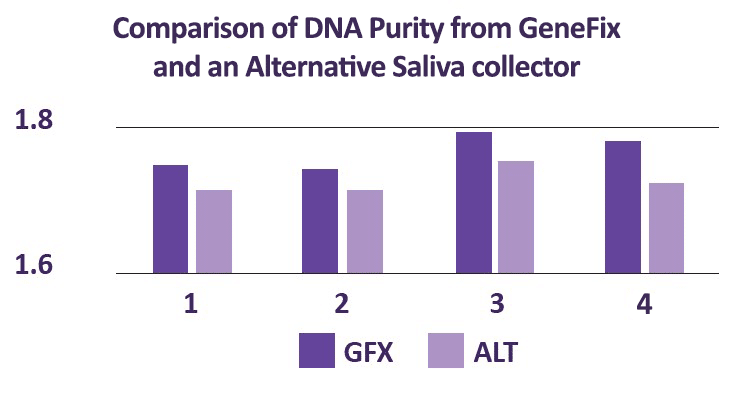
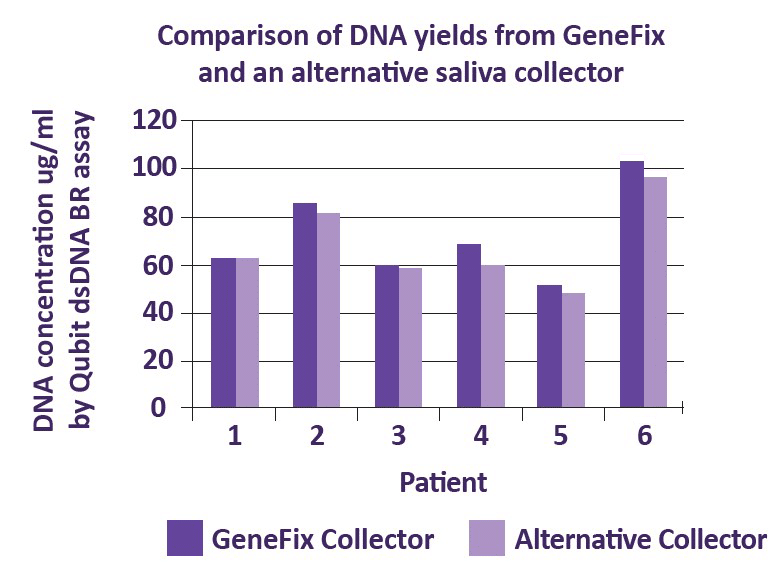
For the oral microbiome, saliva is the best sample type. The GeneFix Saliva Microbiome DNA Collector (MFX) is an easy to use oral microbiome DNA collection kit that has been optimised for the collection of oral microbiome samples using saliva.
Finally, Buccal Swabs (SK) can be used to collect samples from surfaces, either in the environment, or for skin microbiome sampling. Various stabilization options are available to preserve swab samples.
2. Preventing Sample Contamination
Contamination from external sources such as environmental microbes or human DNA, can confound analysis of the gut microbiome. It is important to control for and minimize such contamination to ensure accurate identification and characterization of microbial species present.
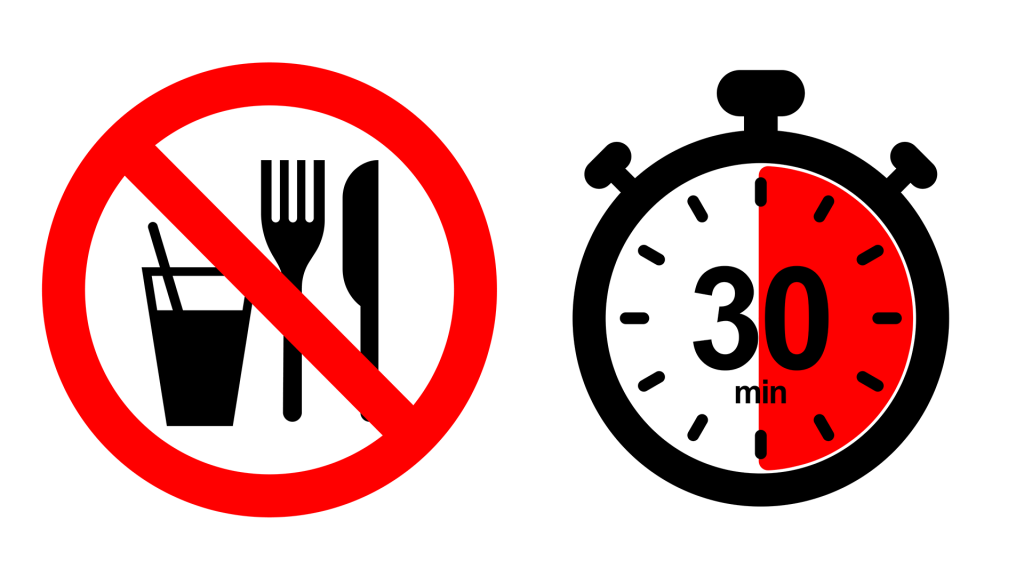
Isohelix collection devices and buffers are free from any traces of human or bacterial DNA, so researchers can be confident in the accuracy of their results. To avoid contamination of oral microbiome samples, patients must avoid eating, drinking, smoking, or brushing their teeth for 30 minutes before sampling.
3. Optimise Extraction
To optimise the extraction of microbial DNA it is important to ensure that collection and stabilization kits are compatible with the microbiome DNA extraction method.
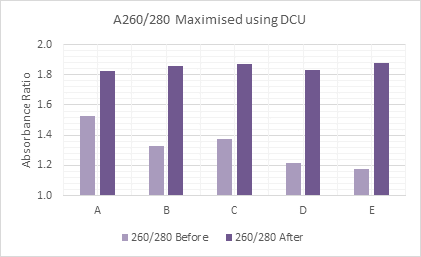
To extract DNA from swabs, for either gut or skin microbiome samples, the BuccalPrep Plus DNA Isolation Kit (BPP) can be used to isolate high quality DNA suitable for all downstream applications. Alternatively, when using Isohelix stabilization buffers such as BuccalFix and StoolFix, BuccalFix Plus DNA Isolation Kit (BFP) can be used to maximize samples.
The Saliva Microbiome DNA collector has been designed for use with the Saliva-Prep2 DNA isolation kit, and produces extremely high yields of microbial DNA (more than twice the yield of other commonly available kits). DNA isolated using Saliva-Prep2 is intact and readily amplifiable, with high purity, and therefore suitable for all downstream analyses.
How to analyse the microbiome
When you have isolated high quality DNA, the two most common methods to measure microbiome depth and diversity are:
- Targeted sequencing of the 16s rRNA gene of bacteria, and internal transcribed spacer (ITS) region sequencing for fungi. These genes are highly conserved, but have diverged over time and can be used to provide a, “barcode” that can be assigned to specific taxonomies, or counted to identify the frequency of each member of the microbial community.
- Shotgun metagenomics – untargeted sequencing methods capture all microbial genomes present within a sample. Metagenomic shotgun assemblies are either performed de novo, based on reference genomes, or a hybrid of both. All types of microorganisms can be sequenced, not just bacteria and fungi.
Tools to study the microbiome are rapidly advancing, and as more researchers begin to investigate the role of the microbiome in health and disease, more effort is spent developing new robust statistical methods and analytical tools required for data analysis.
Future Perspectives
Technical advances in DNA sequencing and bioinformatics analysis have enabled us to understand the importance of microbiomes. Understanding their role, including the dynamic interactions with hosts and their microbes, enables new strategies in a wide range of fields from health to ecology and agriculture.
The manipulation of human microbiota and host-microbiome interactions is a valuable management tool for many health conditions, and some beneficial microbes are even considered next-generation drugs.
At the heart of any microbiome study is a need for high-quality DNA suitable for next-generation sequencing. The Isohelix range of products addresses the challenges with microbiome DNA collection and extraction, enabling researchers to obtain the high-quality DNA they need, to conduct their work.
References
[i] Flint, H.J. The impact of nutrition on the human microbiome. Nutr Rev. Aug;70 Suppl 1:S10-3 (2012).
[ii] Vijay, A., Valdes, A.M. Role of the gut microbiome in chronic diseases: a narrative review. Eur J Clin Nutr 76, 489–501 (2022).
[iii] Sims TT, et. al, Gut microbial diversity and genus-level differences identified in cervical cancer patients versus healthy controls. Gynecol Oncol. Nov;155(2):237-244 (2019)
[iv] Safadi, J.M., Quinton, A.M.G., Lennox, B.R. et al. Gut dysbiosis in severe mental illness and chronic fatigue: a novel trans-diagnostic construct? A systematic review and meta-analysis. Mol Psychiatry 27, 141–153 (2022)
[v] Wei L, Singh R, Ro S, Ghoshal UC. Gut microbiota dysbiosis in functional gastrointestinal disorders: Underpinning the symptoms and pathophysiology. JGH Open Mar 23;5(9):976-987. (2021)
[vi] Dewhirst, F. E. et al. The human oral microbiome. J. Bacteriol. 192, 5002–5017 (2010).
[vii] Highlander, S. K. et al. Deep sequencing of the oral microbiome reveals sig natures of periodontal disease. PLoS ONE 7, https://doi.org/10.1371/journal. pone.0037919 (2012).
[viii] Wilson, B. A. et al. The oral microbiome of early stage Parkinson’s disease and its relationship with functional measures of motor and non-motor function. PLoS ONE 14, https://doi.org/10.1371/journal.pone.0218252 (2019).
[ix] Matsha, T. E. et al. Oral microbiome signatures in diabetes mellitus and periodontal disease. J. Dent. Res. 99, 658–665 (2020).
[x] Buffie, C. G. & Pamer, E. G. Microbiota-mediated colonization resistance against intestinal pathogens. Nat. Rev. Immunol. 13, 790–801 (2013).
[xi] lebba, V. et al. Eubiosis and dysbiosis: the two sides of the microbiota. New Microbiol. 39, 1–12 (2016)
[i] Karpinets, T.V., Wu, X., Solley, T. et al. Metagenomes of rectal swabs in larger, advanced stage cervical cancers have enhanced mucus degrading functionalities and distinct taxonomic structure. BMC Cancer 22, 945 (2022).
[i] GeneFix™ Saliva Collectors & Kits for Human DNA Oral Microbiome Analysis, Application Note: GeneFix™ MFX, December (2018)