As DNA testing becomes more accessible and cost-effective, its applications in research, healthcare, and direct-to-consumer services, such as microbiome and ancestry testing, continue to expand. At Isohelix, we’re proud to support this growth by providing high-quality DNA collection and stabilization solutions. Whether you’re studying rare inherited disorders, using genomics to tailor drug treatments for patients, or helping consumers explore their ancestral roots, the success of your genetic analysis starts with reliable sample collection.
In this article, we examine key application areas where DNA collection plays a pivotal role, ranging from lifestyle genomics to cancer diagnostics. Read on to learn more about:
- Unlocking Complex Traits with GWAS
- Calculating Genetic Health Risks
- Pharmacogenetics and Pharmacogenomics
- Carrier Screening for Inherited Disorders
- Lifestyle genomics
- Microbiomics
Unlocking Complex Traits with GWAS
What is GWAS used for?
Genome-wide association studies (GWAS) compare genetic differences between samples from donors with and without a disease or trait, to find links. The primary genetic variations researchers look for are single nucleotide polymorphisms (SNPs), which represent a difference in a single nucleotide and are the most common type of genetic variation. SNPs commonly occur between genes, where they can be used as biological markers to help scientists locate genes associated with a disease or trait, within a gene, or in a regulatory region near a gene where they may affect the gene function and play a direct role in the trait or disease.
Multi-center GWAS projects often collect DNA from large numbers of individuals, utilizing high-throughput genotyping platforms, such as SNP arrays, to identify associations between SNPs and traits. These studies extract DNA from large cohorts, often over a wide geographical area, that must be collected and preserved to ensure consistent quality across the study.
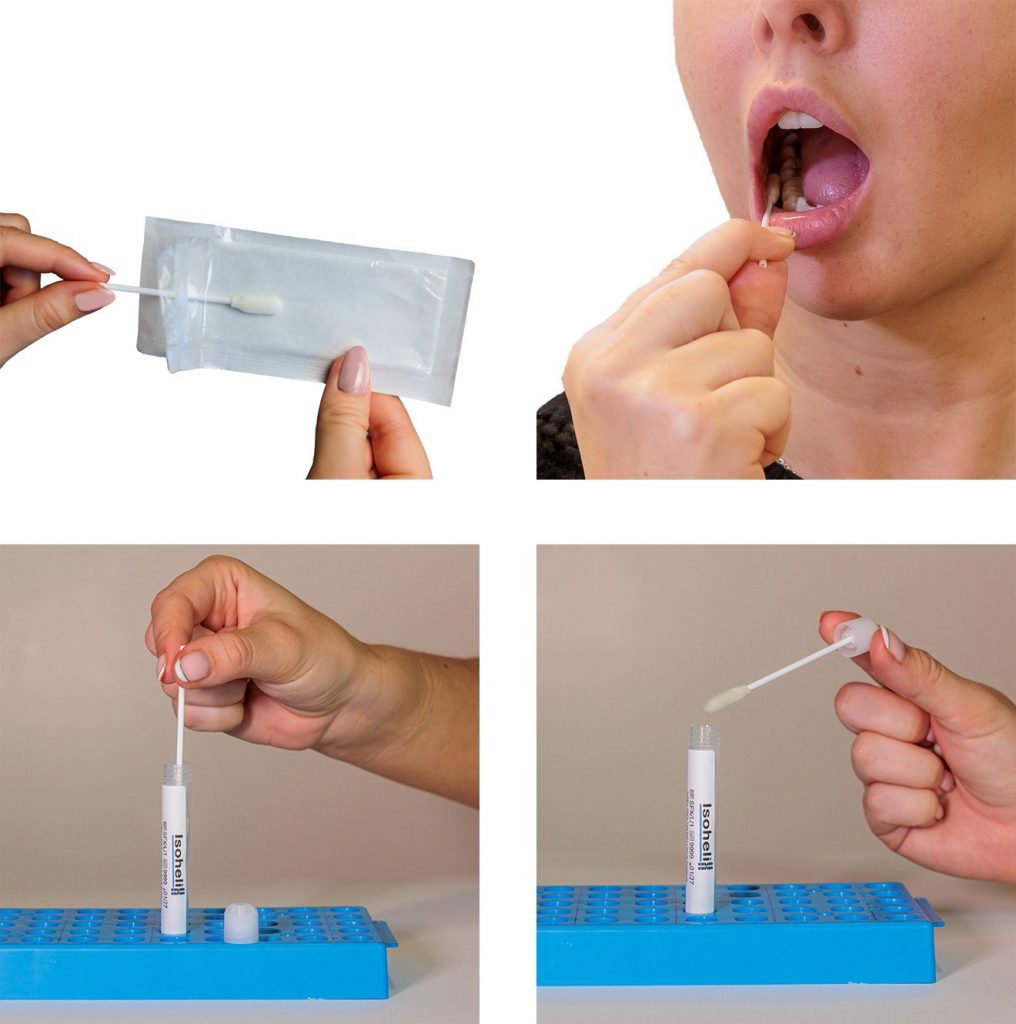
Historically, DNA was collected from blood for GWAS studies, but as DNA collection and stabilization technology have improved for saliva and buccal swabs, these sample types are now commonly used. Saliva collection is a safe and non-invasive process, and saliva collection kits can be mailed to donors for self-collection at home. Read more about how Isohelix saliva collection kits are replacing blood sampling at https://isohelix.com/saliva-instead-of-blood/.
Calculating Genetic Health Risks
Predictive genetic testing enables individuals to understand their risk of developing a specific condition or disease. Identifying genetic risk factors can encourage earlier lifestyle interventions or further clinical screening. A polygenic risk score is used to estimate the genetic risk for a trait or disease by combining the effects of multiple SNPs. Polygenic Risk Scores (PRS) are often generated using genome-wide association study (GWAS) data, which can come from SNP array data generated from DNA samples.
For example, Polygenic Risk Scores (PRS) have been widely used in breast cancer research to stratify risk and inform screening and prevention strategies. Integrating PRS into clinical practice enables healthcare providers to deliver more accurate risk assessments, personalized prevention strategies, and optimized screening programs.
Pharmacogenetics and Pharmacogenomics
What’s the difference between pharmacogenetics and pharmacogenomics? Although the terms are often used interchangeably, pharmacogenetics is a subset of pharmacogenomics, which focuses on understanding how genetic variations influence an individual’s drug response. By contrast, pharmacogenomics refers to the investigation of the collective influence of the entire genome on a drug response.
Pharmacogenetic/genomic testing is revolutionizing precision medicine by helping clinicians prescribe drugs that are more likely to be effective for patients and less likely to cause side effects based on the patient’s genetic makeup.
Knowing a patient’s genotype at a specific locus can help doctors select the correct medication and dosage. For example, variants in the CYP2C19 gene affect how a patient metabolizes clopidogrel, a commonly prescribed blood thinner[i].
Pharmacogenetic/genomic testing is revolutionizing precision medicine by helping clinicians prescribe drugs that are more likely to be effective for patients. and less likely to cause side effects based on the patient’s genetic makeup.
The broader approach of Pharmacogenomics evaluates all genetic interactions, allowing healthcare professionals to design treatment plans that require multiple medications. With the drug Trastuzumab, variations in the HER2 gene[ii], along with other genes that affect drug metabolism and immune function, can impact a patient’s response.
Real-time PCR (qPCR) is often used in pharmacogenetics, where a fast turnaround time is necessary, and a single or small number of genes are being investigated. For pharmacogenomics, whole genome SNP arrays provide genotype information on hundreds of thousands of loci. Blood and/or saliva samples can be used to generate genotyping data with both technologies.
Carrier Screening for Inherited Disorders
Carrier screening identifies individuals who may pass on harmful genetic conditions such as cystic fibrosis, spinal muscular atrophy, or Tay-Sachs disease. Historically, carrier screening was limited to a select number of tests, primarily offered to individuals or populations at higher risk. However, the use of carrier screening and its role in reproductive health is changing rapidly as genetic analysis tools, such as high-density microarrays, next-generation sequencing (NGS), and powerful bioinformatics, become more sophisticated. A single DNA sample can now be used to screen hundreds of disorders.
Lifestyle genomics
From ancestry testing to athletic performance and appearance traits, lifestyle genomic testing has exploded in popularity. Consumers are using at-home DNA collection of saliva or buccal swabs to uncover where their ancestors came from, how their bodies process nutrients, or whether they’re genetically predisposed to faster recovery from exercise. An ancestry company may use SNP genotyping to map thousands of genetic markers, comparing them to global population databases to infer ancestral origins.
Microbiomics
Consumer microbiome testing has rapidly gained popularity in recent years, offering individuals a window into the complex communities of bacteria, fungi, and other microorganisms living in their gut. Using at-home stool collection kits, consumers can submit samples for sequencing and analysis, revealing how their microbiome may influence digestion, immunity, skin health, mood, and even weight management.
Many testing providers pair results with personalized diet or supplement recommendations based on the individual’s microbial profile. The accuracy of microbiome insights relies on high-quality, well-preserved samples, making robust DNA collection and stabilization methods essential to ensure reliable sequencing data.
Isohelix DNA Collection and Stabilization
At Isohelix, we understand the importance of high quality DNA collection, extraction and preservation. Our GeneFixTM collection features high-yielding sample collectors that seamlessly integrate with our expanding range of Isohelix DNA collection, extraction and stabilization kits. These genetic sampling kits are specifically designed to provide exceptional DNA purity and yield, whether for life science research, molecular diagnostics, or forensic applications.
Visit our website to find out more: https://isohelix.com/
[i] Brown SA, Pereira N. Pharmacogenomic Impact of CYP2C19 Variation on Clopidogrel Therapy in Precision Cardiovascular Medicine. J Pers Med. 2018 Jan 30;8(1):8. doi: 10.3390/jpm8010008. PMID: 29385765; PMCID: PMC5872082.
[ii] Zakaria NH, Hashad D, Saied MH, Hegazy N, Elkayal A, Tayae E. Genetic mutations in HER2-positive breast cancer: possible association with response to trastuzumab therapy. Hum Genomics. 2023 May 18;17(1):43. doi: 10.1186/s40246-023-00493-5. PMID: 37202799; PMCID: PMC10193616.
Further Reading
- Genome Wide Association Studies (GWAS)
Milona, M., et al. Association of Three Genetic Loci with Molar Incisor Hypomineralization in Polish Children. J. Clin. Med. 2024, 13, 857. https://doi.org/10.3390/jcm13030857
Mitchell, Brittany L., et al. “Polygenic risk scores derived from varying definitions of depression and risk of depression.” JAMA psychiatry 78.10 (2021): 1152-1160. https://doi.org/10.1001/jamapsychiatry.2021.1988
Lind, Penelope A., et al. “Clozapine efficacy and adverse drug reactions among a nationwide study of 1021 Australians prescribed clozapine: The ClozaGene Study.” Schizophrenia Bulletin (2024): sbae065. https://doi.org/10.1093/schbul/sbae065
Rajan-Babu, Indhu-Shree, et al. “Defining the performance parameters of a rapid screening tool for FMR1 CGG-repeat expansions based on direct triplet-primed PCR and melt curve analysis.” The Journal of Molecular Diagnostics 18.5 (2016): 719-730. https://doi.org/10.1016/j.jmoldx.2016.05.002
Kazan, H.H., Bulgay, C., Zorba, E. et al. Exploring the relationship between caffeine metabolism-related CYP1A2 rs762551 polymorphism and team sport athlete status and training adaptations. Mol Biol Rep 51, 841 (2024). https://doi.org/10.1007/s11033-024-09800-2
Saifon, W., Sensorn, I., Trachu, N. et al. Gastrointestinal microbiota profile and clinical correlations in advanced EGFR-WT and EGFR-mutant non-small cell lung cancer. BMC Cancer 22, 963 (2022). https://doi.org/10.1186/s12885-022-10050-3