Blood samples have traditionally been considered the gold standard DNA source for genomic analysis, but obtaining blood samples is a painful, invasive, and costly procedure that must be performed by qualified personnel.
By contrast, saliva collection is safe and non-invasive, and saliva collection kits can be mailed to donors for self-collection at home. As well as the sample source, how the sample is collected, stabilized, stored, and purified is critical to the quality and quantity of DNA that can be extracted.
Previously, challenges with DNA extracted from saliva included microbial contamination and lower nucleic acid yields. However, there have been significant improvements in devices to collect saliva, and good yields of high-quality DNA can now be extracted
This article discusses the use of DNA extracted from saliva for a wide range of downstream applications
Saliva is much easier and cheaper to collect and transport than blood
Participants in large scale epidemiological studies may be reluctant to provide blood samples due to the need to travel to a health center or the painful nature of giving a sample. Recruitment and compliance rates are much higher when saliva samples are used, as saliva collection is painless and can done at home.
Traditionally blood samples were shipped and stored at -20 degrees before processing, but DNA stabilization reagents are now available for collecting, transporting, and storing whole blood. However, these reagents often contain hazardous substances such as guanidium, so care must be taken while handling them.
Saliva samples require no pre-processing and are commonly collected using non-hazardous reagents, so they can be sent via mail. Saliva collection kits can stabilize DNA at room temperature for over five years, avoiding the high cost and logistical challenges of cold chain transport.
Which cells provide the DNA extracted from saliva and blood samples?
High quality DNA extracted from peripheral blood originates from leukocytes.
Human DNA in saliva originates from epithelial cells or or leukocytes. Unlike DNA derived from blood, saliva samples can include bacterial DNA, allowing DNA extraction from the oral microbiome. If required, qPCR assays can be used to quantify either human or bacterial DNA and measure and normalize saliva DNA samples.
Isohelix estimated the relative quantities of human and microbial DNA found in samples stabilized using their GeneFix reagent, which were then stored at room temperature prior to DNA extraction [i]. Approximately 7.4% of DNA in the samples was found to be of microbial origin. In a follow up study {ii}, GeneFix saliva stability samples that had been stored for up to 48 months at room temperature were tested, and no change in the proportion of bacterial DNA in samples over the stability period was found, demonstrating the GeneFix collectors fully stabilize saliva samples, preserve DNA and prevent microbial growth.
DNA yield from saliva samples is comparable to blood for downstream applications
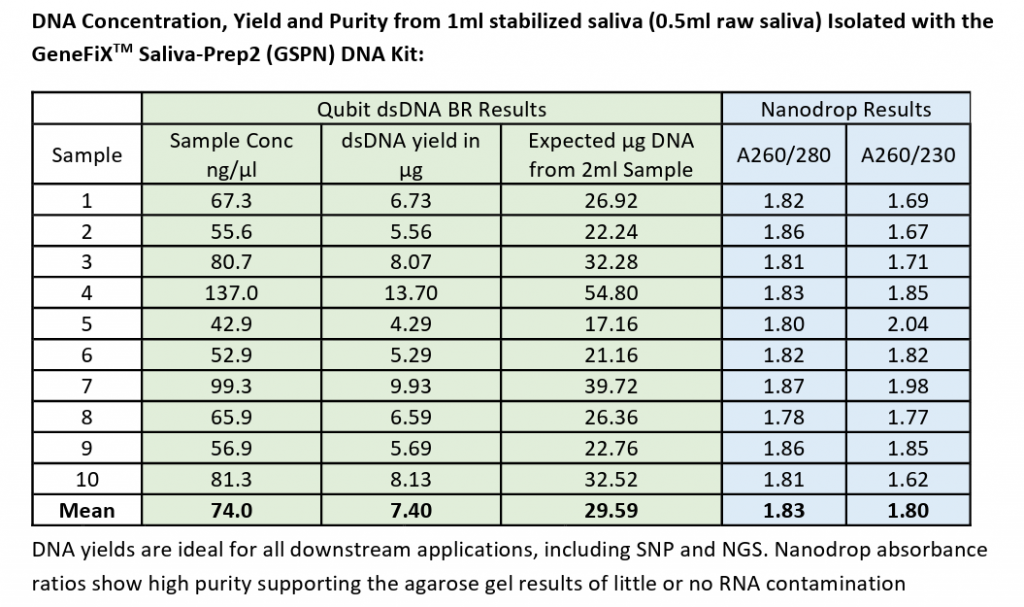
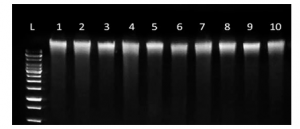
Figure 1 shows the DNA concentration, yield and purity from 0.5ml raw saliva isolated using the GeneFix Saliva-Prep2 DNA Kit [iii}
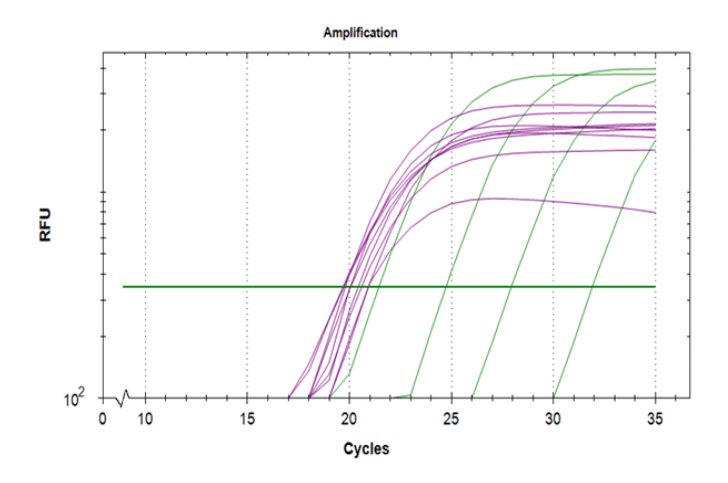
In a study comparing DNA extracted from saliva and blood, Looi et al [iv] (2012), the DNA yield from saliva of 7.8 µg/0.5 mL from a manual purification method was comparable to the DNA yield from blood using a salt precipitation method (7.4 µg/0.5 mL blood sample). DNA extracted from saliva and blood were both high purity (A260/280 > 1.70).
Downstream analysis using DNA from saliva
The quality and quantity of DNA required for a study depend on the downstream analysis that will be performed. Below are some recent studies demonstrating the successful use of DNA from saliva extracted using Isohelix kits. For a more comprehensive list of publications please click here
PCR and RT-PCR
DNA extracted from saliva is routinely used for PCR and qRT-PCR and became particularly important during the Covid 19 pandemic.
Some key examples are :
- Potocka, Natalia, et al. “Association of ACTN3 Polymorphism with Body Somatotype and Cardiorespiratory Fitness in Young Healthy Adults.” International journal of environmental research and public health 16.9 (2019): 1489. https://doi.org/10.3390/ijerph16091489
- Carter, Nikki, et al. “A novel automated SARS-CoV-2 saliva PCR test protects a global asymptomatic workforce.” Scientific Reports 11.1 (2021): 1-6. https://doi.org/10.1038/s41598-021-92070-w
Genotyping
The accuracy of genotyping with saliva-derived DNA has been reported as comparable to DNA derived from blood [v,vi]
There are many examples in the literature where saliva samples have been used for genotyping :
- Campos, Adrian I., et al. “Impact of CYP2C19 metaboliser status on SSRI response: a retrospective study of 9500 participants of the Australian Genetics of Depression Study.” The Pharmacogenomics Journal 22.2 (2022): 130-135. https://doi.org/10.1038/s41397-022-00267-7
- Potocka, Natalia, et al. “Effects of the Trp64Arg Polymorphism in the ADRB3 Gene on Body Composition, Cardiorespiratory Fitness, and Physical Activity in Healthy Adults.” Genes8 (2023): 1541. https://doi.org/10.3390/genes14081541
Next Generation Sequencing
Although the use of blood-derived DNA is the current standard for WGS, Wall et al [vii] reported no differences in sequencing quality or variant call error rate between blood and saliva samples for both whole exome sequencing (WES) and WGS.
There are several examples in the literature of DNA from saliva being used for NGS :
- Hansen, Marcus Høy, and Charlotte Guldborg Nyvold. “Replicate whole-genome next-generation sequencing data derived from Caucasian donor saliva samples.” Data in Brief 38 (2021): 107349. https://doi.org/10.1016/j.dib.2021.107349
- Gopinath, Divya, et al. “Salivary bacterial shifts in oral leukoplakia resemble the dysbiotic oral cancer bacteriome.” Journal of oral microbiology 13.1 (2021): 1857998. https://doi.org/10.1080/20002297.2020.1857998
Methylation-based studies
There are some challenges with using DNA extracted from saliva for methylation-based analyses, e.g., cellular heterogeneity in salivary DNA, saliva samples can include bacterial DNA, and saliva samples are fragmented making long-range PCR or long read sequencing difficult [viii].
However, there are several examples in the literature of DNA from saliva being used for methylation studies, e.g., :
- Ruffell, Simon GD, et al. “Ceremonial Ayahuasca in Amazonian Retreats—Mental Health and Epigenetic Outcomes From a Six-Month Naturalistic Study.” Frontiers in Psychiatry 12 (2021): 898. https://doi.org/10.3389/fpsyt.2021.687615
- Zhang, Jun, et al. “Exploring Effect of Postdischarge Developmental Support Program on Preterm Infant Neurodevelopment and BDNF Gene DNA Methylation.” Advances in Neonatal Care (2022): 10-1097i
Summary
Saliva collection is a robust, non-invasive and low cost method of gathering samples that is particularly useful for large-scale epidemiological and other genetic studies where recruitment rates are much higher when saliva samples are used, rather than blood.
Saliva DNA can be used for a wide range of analyses, including PCR, RTPCR, Genotyping Arrays, and Next Generation Sequencing.
References
[i] “Existing Human and Bacterial DNA Content in Human Saliva Samples,” March 2023
[iv] Looi ML, Zakaria H, Osman J, Jamal R. Quantity and quality assessment of DNA extracted from saliva and blood. Clin Lab. 2012;58(3-4):307-12. PMID: 22582505
[v] Genetic epidemiology : Ng DP, Koh D, Choo S, Chia KS. Saliva as a viable alternative source of human genomic DNA in genetic epidemiology. Clin Chim Acta. 2006;367(1–2):81–5.
[vi] Gudiseva HV, Hansen M, Gutierrez L, Collins DW, He J, Verkuil LD, et al. Saliva DNA quality and genotyping efficiency in a predominantly elderly population. BMC Med. Genom. [Internet]. (2016). 2016 Apr 7
[vii] Wall JD, Tang LF, Zerbe B, Kvale MN, Kwok PY, Schaefer C, et al. Estimating genotype error rates from high-coverage next-generation sequence data. Genome Res. 2014;24(11):1734–9
[viii] Nishitani S, Parets SE, Haas BW, Smith AK. DNA methylation analysis from saliva samples for epidemiological studies. Epigenetics. 2018;13(4):352-362. doi: 10.1080/15592294.2018.1461295. Epub 2018 Aug 1. PMID: 29912612; PMCID: PMC6140812


 Email Us
Email Us